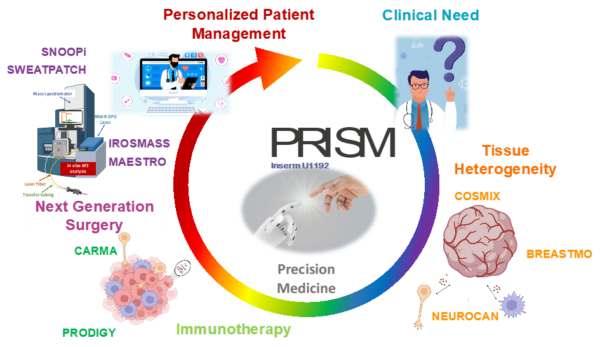
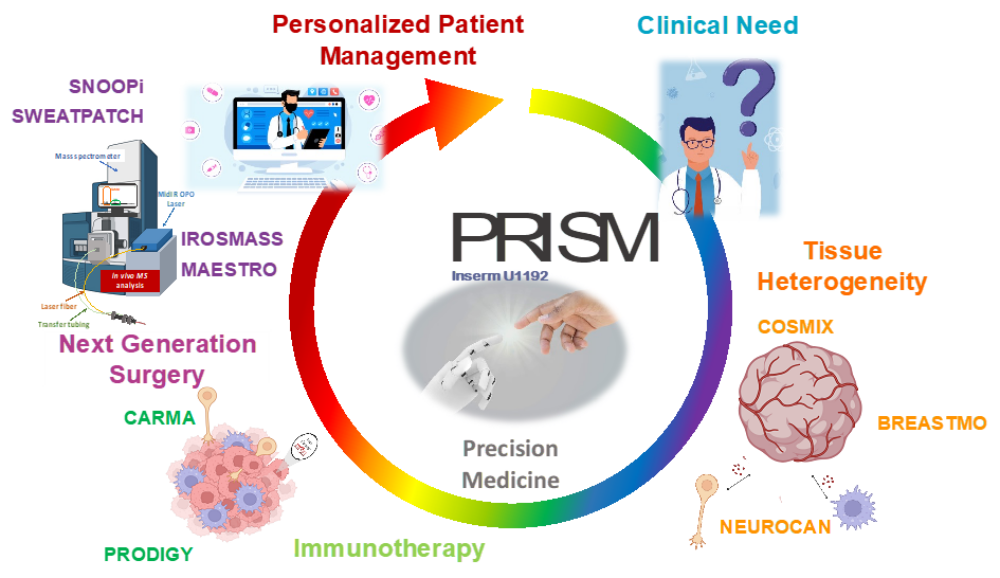
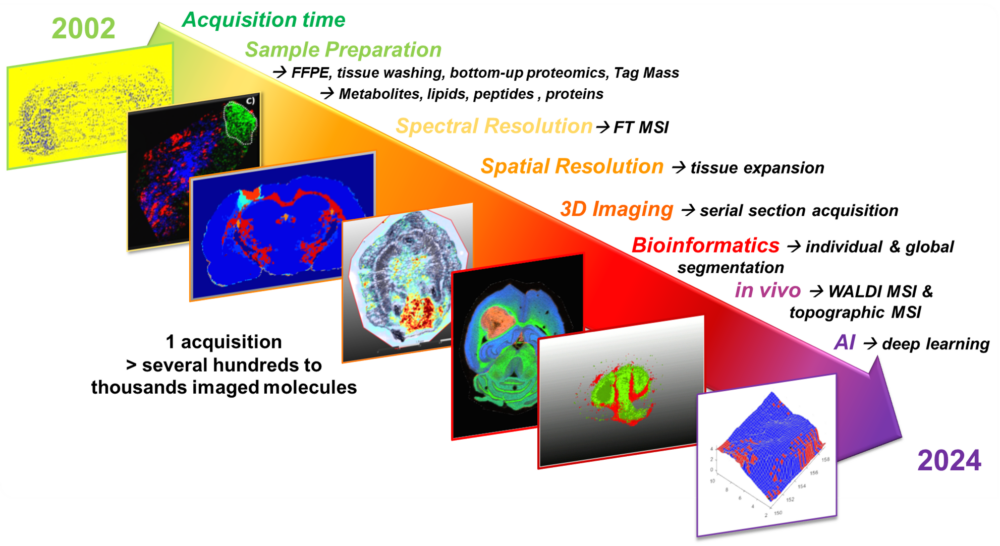
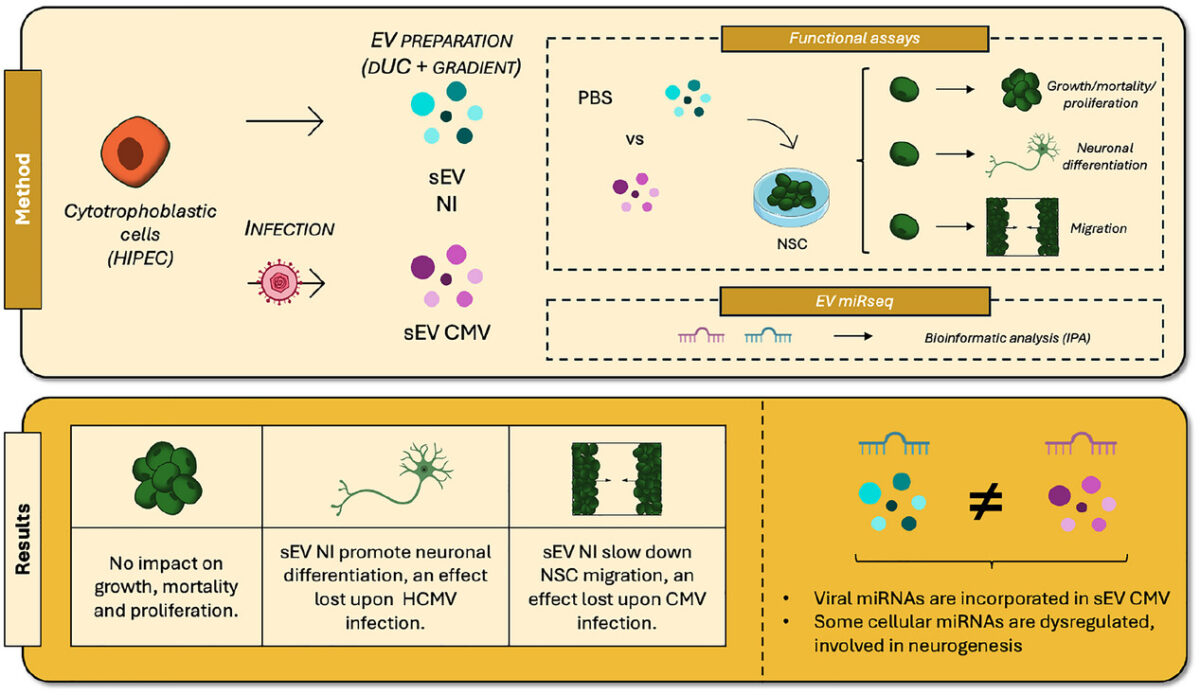
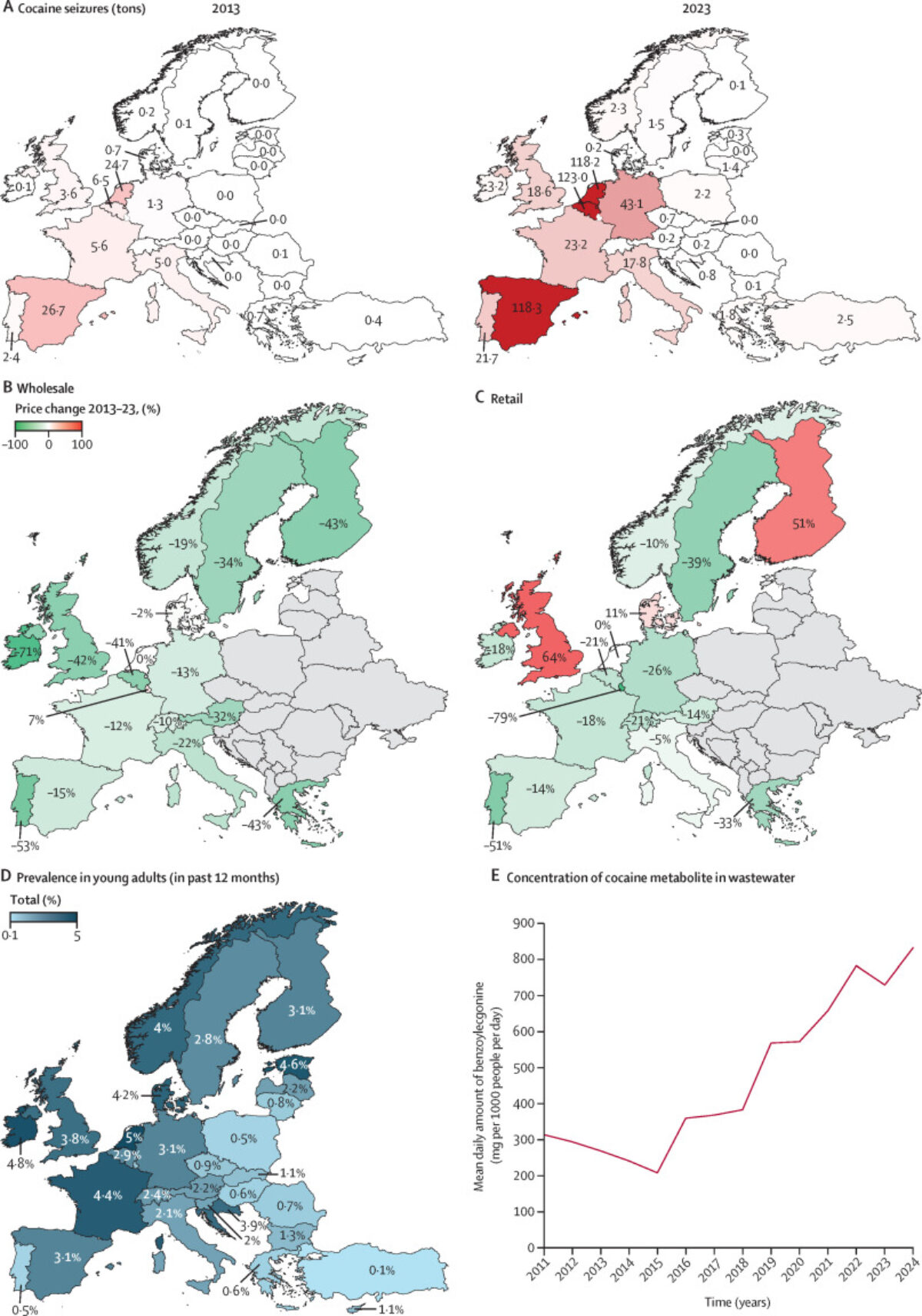
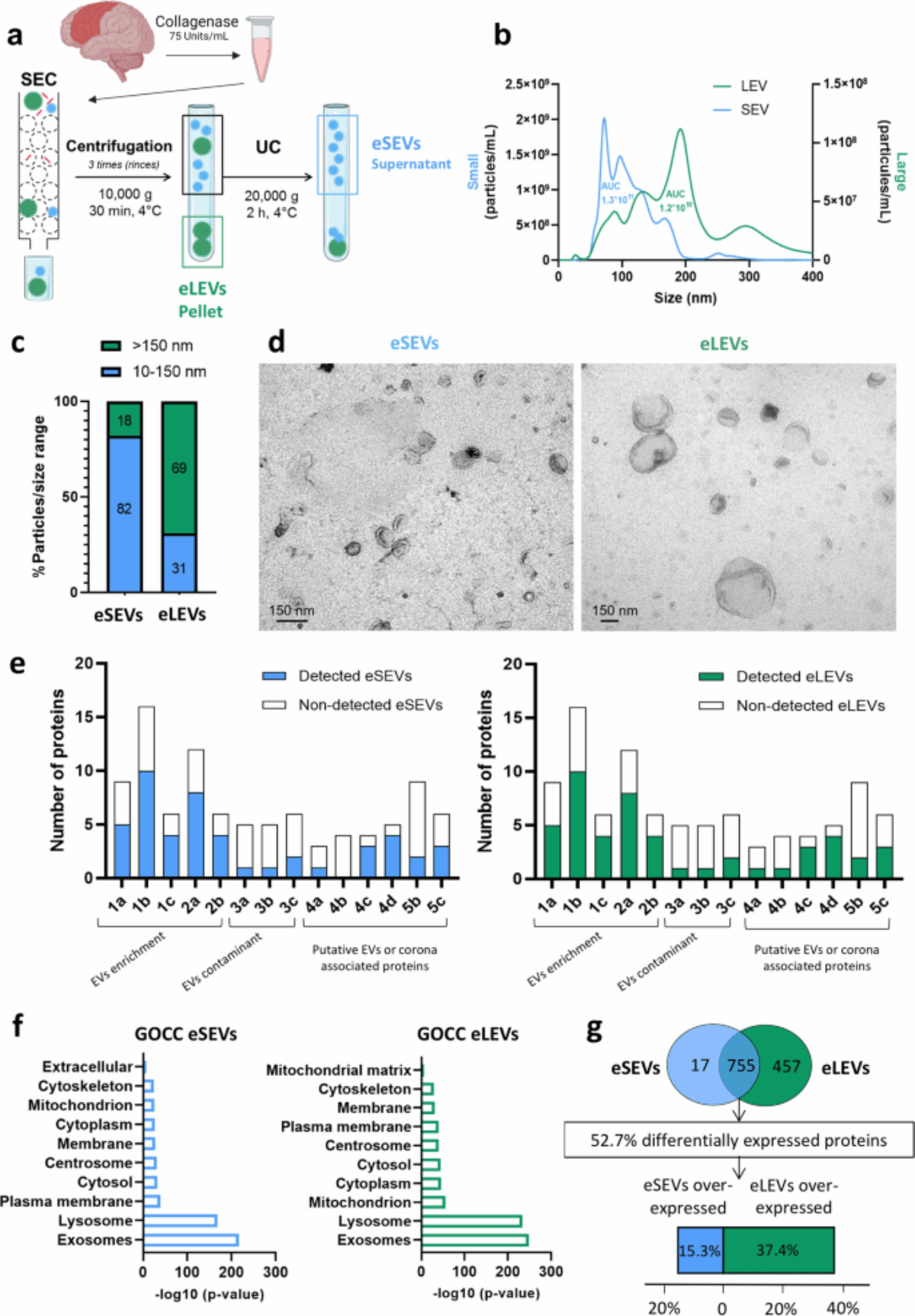
At PRISM, we are driven by a bold vision: to revolutionize healthcare by developing cutting-edge technological and therapeutic innovations that address critical clinical challenges. As an interdisciplinary research unit, we bring together experts in mass spectrometry, spatial proteomics, immunotherapy, and biomedical engineering to push the boundaries of medical science.
Our research is structured around two key pillars:
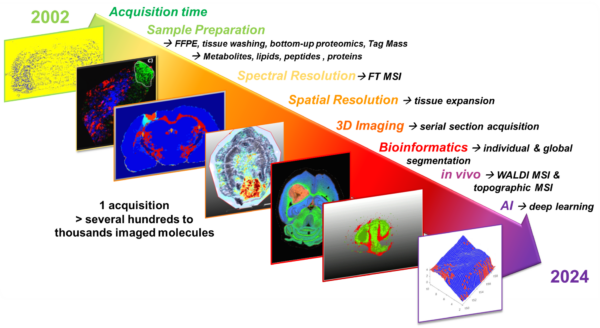
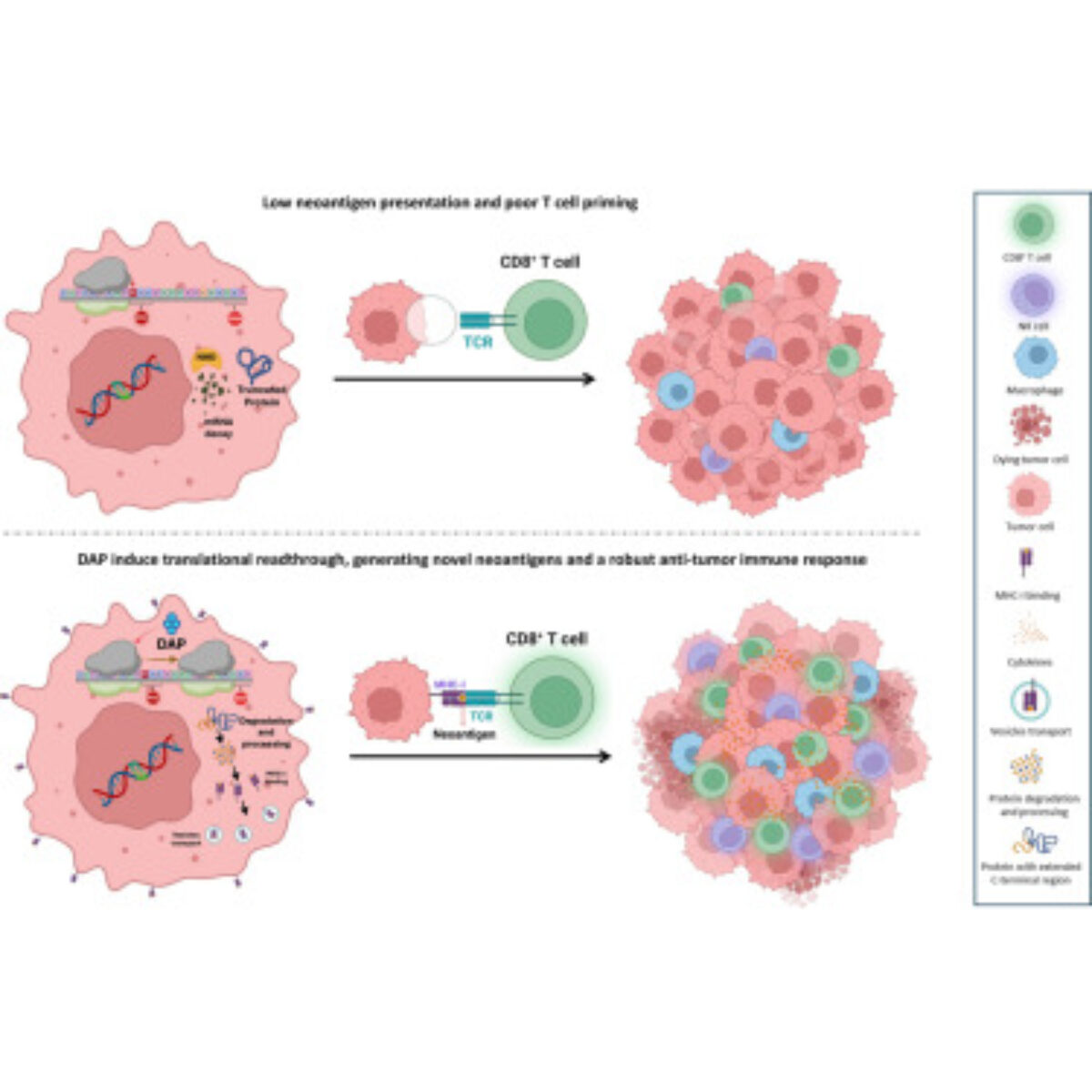
- The Technological Innovation– We develop state-of-the-art technologies to explore the complexity of tissues at the molecular level. By combining single-cell and spatial proteomics and minimally invasive surgical tools, we aim to uncover new disease markers and stratify patients for more precise and effective treatments.
- The Therapeutic innovations – We translate our discoveries into novel therapies, from next-generation cancer immunotherapies. Our goal is to bring groundbreaking solutions from the lab to the clinic, improving patient care through personalized medicine.
Through strong partnerships with clinical centers such as CLCC Oscar Lambret and CHU Lille, as well as industry collaborations, PRISM is at the forefront of translational research. We are committed to transforming fundamental scientific discoveries into real-world applications, pioneering new diagnostic tools, advanced surgical techniques, and breakthrough immunotherapies.
With a relentless pursuit of innovation, PRISM is shaping the future of biomedical research—where technology meets therapy to redefine patient outcomes.